Don't have an account? Register!
Please Login to Use Workflows
Checkout the upcoming new GNPS homepage here!
Data Analysis
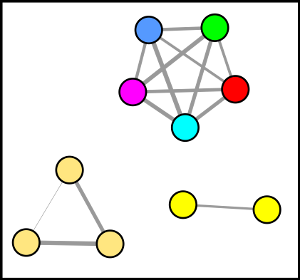 The Data Analysis portal will allow you to organize and visualize your mass spectrometry data. Leveraging the molecular networking techniques, there are additional tools to aid in understanding the unknowns in your sample.
Check out the documentation and live demo.
Further, a separate dereplication workflow is provided as a standalone workflow.
The Data Analysis portal will allow you to organize and visualize your mass spectrometry data. Leveraging the molecular networking techniques, there are additional tools to aid in understanding the unknowns in your sample.
Check out the documentation and live demo.
Further, a separate dereplication workflow is provided as a standalone workflow.
Create Public MassIVE Datasets
 Submit your own data to be made public MassIVE datasets. These MassIVE datasets must be prefixed with GNPS to be visible to other GNPS users. Take advantage of continuous identification to learn more about your dataset after publication automatically. New hits to the community curated libraries and related datasets are reported. Documentation
Submit your own data to be made public MassIVE datasets. These MassIVE datasets must be prefixed with GNPS to be visible to other GNPS users. Take advantage of continuous identification to learn more about your dataset after publication automatically. New hits to the community curated libraries and related datasets are reported. Documentation
Contribute to Libraries
 Be a part of the collaborative community effort to create the definitive collection of natural products MS/MS spectra.
Additionally users can contribute varying levels of quality of spectra: bronze, silver, gold.
GNPS gives the power to add spectra, update annotations, and facilitate dialog around these spectra, to provide a truly collaborative and open natural product MS/MS database.
For documentation and definition of quality requirements click here.
To make corrections to and comments on exisiting spectra in the libraries, users should refer to this documentation.
Be a part of the collaborative community effort to create the definitive collection of natural products MS/MS spectra.
Additionally users can contribute varying levels of quality of spectra: bronze, silver, gold.
GNPS gives the power to add spectra, update annotations, and facilitate dialog around these spectra, to provide a truly collaborative and open natural product MS/MS database.
For documentation and definition of quality requirements click here.
To make corrections to and comments on exisiting spectra in the libraries, users should refer to this documentation.
MassIVE Public GNPS Datasets
 Browse publically available datasets. Here you can download these datasets as well as comment on them so others in the community can see any updates or any new analysis.
Additionally, users can subscribe to the datasets and get updates when new identifications are made via GNPS's continuous identification.
To read further on how to take advantage of the subscriptions to MassIVE datasets and other social networking features click here .
Browse publically available datasets. Here you can download these datasets as well as comment on them so others in the community can see any updates or any new analysis.
Additionally, users can subscribe to the datasets and get updates when new identifications are made via GNPS's continuous identification.
To read further on how to take advantage of the subscriptions to MassIVE datasets and other social networking features click here .
Browse Community Spectral Library
 Browse the community contributed and community curated spectral libraries of natural products. These MS/MS libraries are community contributed and community curated. Users can peak at the inside of these libraries, as well as use them for data analysis. If corrections need to be made, users should refer to this documentation.
Browse the community contributed and community curated spectral libraries of natural products. These MS/MS libraries are community contributed and community curated. Users can peak at the inside of these libraries, as well as use them for data analysis. If corrections need to be made, users should refer to this documentation.
Molecular Explorer
 Bridge the connection between molecules and datasets. Explore exactly where certain molecules are found in all the publically available dataset at GNPS. Powered by GNPS's continuous identification, users are able to see not only which datasets contain what compound, but how many known and unknown analogs exist in all datasets!
Bridge the connection between molecules and datasets. Explore exactly where certain molecules are found in all the publically available dataset at GNPS. Powered by GNPS's continuous identification, users are able to see not only which datasets contain what compound, but how many known and unknown analogs exist in all datasets!
Rarefaction Curve Generation
 The Data Analysis portal will additionally provide a method to create rarefaction curves. These curves will allow you to assess the diversity of MS/MS spectra and ultimately of compounds within your data. For documentation click here.
The Data Analysis portal will additionally provide a method to create rarefaction curves. These curves will allow you to assess the diversity of MS/MS spectra and ultimately of compounds within your data. For documentation click here.
Challenge Spectra
 There will always be spectra that stumps the best. Scientists can share these intriguing unidentified yet interesting spectra as challenge spectra.
To deposit challenge spectra click here and please refer to the documentation. To browse user uploaded challenge spectra, click here.
There will always be spectra that stumps the best. Scientists can share these intriguing unidentified yet interesting spectra as challenge spectra.
To deposit challenge spectra click here and please refer to the documentation. To browse user uploaded challenge spectra, click here.
GNPS Theoretical/Insilico
All things related to theoretical/insilico dereplication of natural products. Browse the tools and theoretical MS/MS libraries avaiable at GNPS.
Leverage GNPS Libraries and Datasets
Find where your MS/MS spectrum occurs in all GNPS datasets. Check it out!
Documentation
GNPS Experimental
Browse experimental tools at GNPS.
Citation
Wang, Mingxun, et al. "Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking." Nature Biotechnology 34.8 (2016): 828-837. PMID: 27504778
Copyright © 2025.
Last modified: 2025-01-22.
Version 1.3.16-GNPS.
